class: middle, left, title-slide # Putting the <img src='assets/logos/R_logo.svg' height=45px> into Reproducible Research ## <br/> ### <strong>Dr Anna Krystalli </strong> <br/> University of Sheffield RSE ### <br> 2019/05/14 - <small> RSS Sheffield Local Group meeting </small> --- layout: true <div class="my-footer"><span>RSS Sheffield Local Group meeting                             @annakrystalli <a href="https://twitter.com/annakrystalli"><i class="fab fa-twitter"></i> </a> @annakrystalli <a href="https://github.com/annakrystalli"><i class="fab fa-github-alt"></i> </a> </span></div> --- # 👋 Hello ### me: **Dr Anna Krystalli** - **Research Software Engineer**, _University of Sheffield_ + twitter **@annakrystalli** + github **@annakrystalli** + email **a.krystalli[at]sheffield.ac.uk** - **Editor [rOpenSci](http://onboarding.ropensci.org/)** - **Co-organiser:** [Sheffield R Users group](https://www.meetup.com/SheffieldR-Sheffield-R-Users-Group/) <br> ### slides: **bit.ly/r-in-repro-research** --- class: inverse, center, middle # Motivation --- # Calls for reproducibility > ### **Reproducibility** has the potential to serve as a **minimum standard for judging scientific claims** when full independent replication of a study is not possible. <br> .center[ <img src="assets/repro-spectrum.jpg" width="650px" /> ] <br> --- # Is code and data enough? .center[  ] .img-attr[slide: [_Karthik Ram: rstudio::conf 2019 talk_](https://github.com/karthik/rstudio2019)] --- # Calls for open science <img src="assets/pgls.png" width="350px" href="https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4957270/"/> > ... highlight problems with users jumping straight into software implementations of methods (e.g. in r) that may lack documentation on biases and assumptions that are mentioned in the original papers. > <small> To help solve these problems, we make a number of suggestions including **providing blog posts** or **videos** to **explain new methods in less technical terms**, **encouraging reproducibility and code sharing**, making **wiki-style pages** summarising the literature on popular methods, more careful consideration and testing of whether a method is appropriate for a given question/data set, **increased collaboration**, and a shift from publishing purely novel methods to **publishing improvements to existing methods** and ways of detecting biases or testing model fit. Many of these points are applicable across methods in ecology and evolution, not just phylogenetic comparative methods.</small> --- # R for Open Reproducible Research ## A whistle-stop tour of tools, practices and conventions in R for more: -- - ### **Reproducible** -- - ### **Robust** -- - ### **Transparent** -- - ### **Reusable** -- - ### **Shareable** research materials --- class: inverse, center, middle # Project management <img src="assets/project.svg" height=200px> .img-attr.bottom[Icon by [Freepik](https://www.freepik.com/) from [flaticon.com](www.flaticon.com)] --- # Rstudio Projects ## Use Rstudio projects to keep materials associated with a particular analysis together <br> .pull-left[ - **Self contained** and **portable** - **Working directory set to root** of project on launch - **Fresh session** everytime the project is launched See Jenny Bryan's post on [**project oriented workflows**](https://www.tidyverse.org/articles/2017/12/workflow-vs-script/) for more details ] .pull-right.center[ **File > New Project > New Directory** <img src="assets/new_project.png" height=200px> ] --- background-image: url('assets/my_awesome_project.png') background-size: contain --- # 📦 `here` ## Use 📦 `here` to create robust relative paths <br> .pull-left[ - **Robust paths relative to project root** + portable + independent of: - working directory - source code location ] .pull-right.center[ <img src="assets/you-are-here.svg" height=150px> ] ```r here::here() ``` ``` ## [1] "/Users/Anna/Documents/workflows/talks" ``` ```r here::here("data", "summaries.csv") ``` ``` ## [1] "/Users/Anna/Documents/workflows/talks/data/summaries.csv" ``` .img-attr.right[Icon by [Freepik](https://www.freepik.com/) from [flaticon.com](www.flaticon.com)] --- # Dependency management ### Minimal approach ##### include an `install.R` script ```r install.packages("dplyr") install.packages("purrr") ``` .pull-left[ ### Most robust approach ##### use 📦 `packrat` - Create and manage a per project library of packages - intialise during project set up ] .pull-right[ <img src="assets/packrat_init.png" height=200px> ] _will revisit later on_ --- # 📦 `drake` ## Use 📦 `drake` to orchestrate your workflows <br> .pull-left[ <img src="https://camo.githubusercontent.com/e1e4b1e1480ab4935b20edfcc4dfa06f0796d993/68747470733a2f2f726f70656e7363692e6769746875622e696f2f6472616b652f666967757265732f696e666f677261706869632e737667" height=150px> ] .pull-right.center[ <img src="https://camo.githubusercontent.com/f14e18db7b61e6f9adbc51fe3fa87893d037f82e/68747470733a2f2f726f70656e7363692e6769746875622e696f2f6472616b652f666967757265732f6c6f676f2e737667" height=150px> ] #### make plan ```r plan <- drake::drake_plan( raw_data = readr::read_csv(here::here("data", "iris.csv")), data = raw_data %>% dplyr::mutate(Species = forcats::fct_inorder(Species)), fit = lm(Sepal.Width ~ Petal.Width + Species, data)) ``` --- # Plan #### view plan ```r plan ``` ``` ## # A tibble: 3 x 2 ## target command ## <chr> <expr_lst> ## 1 raw_data readr::read_csv(here::here("data", "iris.csv")) … ## 2 data raw_data %>% dplyr::mutate(Species = forcats::fct_inorder(Speci… ## 3 fit lm(Sepal.Width ~ Petal.Width + Species, data) … ``` #### re-execute plan ```r drake::make(plan) ``` ``` ## All targets are already up to date. ``` --- #### inspect targets ```r drake::readd(fit) ``` ``` ## ## Call: ## lm(formula = Sepal.Width ~ Petal.Width + Species, data = data) ## ## Coefficients: ## (Intercept) Petal.Width Speciesversicolor ## 3.236 0.781 -1.501 ## Speciesvirginica ## -1.844 ``` --- # visualise workflow .centre[ ```r drake::vis_drake_graph(drake::drake_config(plan)) ``` <div id="htmlwidget-f5734015a6c70f557035" style="width:864px;height:360px;" class="visNetwork html-widget"></div> <script type="application/json" data-for="htmlwidget-f5734015a6c70f557035">{"x":{"nodes":{"id":["n-NBSXEZJ2HJUGK4TF","n-OJSWCZDSHI5HEZLBMRPWG43W","n-MZXXEY3BORZTUOTGMN2F62LON5ZGIZLS","n-MRYGY6LSHI5G25LUMF2GK","raw_data","data","fit"],"imported":[true,true,true,true,false,false,false],"label":["here::here","readr::read_csv","forcats::fct_inorder","dplyr::mutate","raw_data\n0.07s","data\n0.013s","fit\n0.011s"],"status":["imported","imported","imported","imported","up to date","up to date","up to date"],"type":["function","function","function","function","object","object","object"],"font.size":[20,20,20,20,20,20,20],"color":["#1874CD","#1874CD","#1874CD","#1874CD","#228B22","#228B22","#228B22"],"shape":["triangle","triangle","triangle","triangle","dot","dot","dot"],"level":[1,1,1,1,2,3,4],"x":[-1,-1,-1,-1,-0.333333333333333,0.333333333333333,1],"y":[-1,-0.333333333333333,0.333333333333333,1,-2.22044604925031e-16,-2.22044604925031e-16,-2.22044604925031e-16]},"edges":{"from":["n-NBSXEZJ2HJUGK4TF","n-OJSWCZDSHI5HEZLBMRPWG43W","raw_data","n-MZXXEY3BORZTUOTGMN2F62LON5ZGIZLS","n-MRYGY6LSHI5G25LUMF2GK","data"],"to":["raw_data","raw_data","data","data","data","fit"],"arrows":["to","to","to","to","to","to"]},"nodesToDataframe":true,"edgesToDataframe":true,"options":{"width":"100%","height":"100%","nodes":{"shape":"dot","physics":false},"manipulation":{"enabled":false},"layout":{"hierarchical":{"enabled":true,"direction":"LR"}},"interaction":{"navigationButtons":true}},"groups":null,"width":null,"height":null,"idselection":{"enabled":false,"style":"width: 150px; height: 26px","useLabels":true,"main":"Select by id"},"byselection":{"enabled":false,"style":"width: 150px; height: 26px","multiple":false,"hideColor":"rgba(200,200,200,0.5)"},"main":{"text":"Dependency graph","style":"font-family:Georgia, Times New Roman, Times, serif;font-weight:bold;font-size:20px;text-align:center;"},"submain":null,"footer":null,"background":"rgba(0, 0, 0, 0)","highlight":{"enabled":false,"hoverNearest":false,"degree":1,"algorithm":"all","hideColor":"rgba(200,200,200,0.5)","labelOnly":true},"collapse":{"enabled":true,"fit":false,"resetHighlight":true,"clusterOptions":null,"keepCoord":true,"labelSuffix":"(cluster)"},"legend":{"width":0.2,"useGroups":false,"position":"left","ncol":1,"stepX":100,"stepY":100,"zoom":true,"nodes":{"label":["Up to date","Imported","Object","Function"],"color":["#228B22","#1874CD","#888888","#888888"],"shape":["dot","dot","dot","triangle"],"font.color":["black","black","black","black"],"font.size":[20,20,20,20],"id":[1,5,7,8]},"nodesToDataframe":true},"tooltipStay":300,"tooltipStyle":"position: fixed;visibility:hidden;padding: 5px;white-space: nowrap;font-family: verdana;font-size:14px;font-color:#000000;background-color: #f5f4ed;-moz-border-radius: 3px;-webkit-border-radius: 3px;border-radius: 3px;border: 1px solid #808074;box-shadow: 3px 3px 10px rgba(0, 0, 0, 0.2);"},"evals":[],"jsHooks":[]}</script> ] --- class: inverse, center, middle # Version Control --- # Version Control ### What is it? 🤔 The **management of changes** to documents, computer programs, large web sites, and other collections of information. ### Git <img src="https://git-scm.com/images/logos/downloads/Git-Logo-2Color.png" height="25px" > Open source (free to use) **Version control software.** ### GitHub <img src="https://raw.githubusercontent.com/annakrystalli/rrresearch/master/docs/assets/github_logo.jpg" height="25px"> A **website** (https://github.com/) that allows you to **store your Git repositories online** and makes it easy to collaborate with others. --- # Why use them in research? .pull-left[ ### Exhibit A <img src="http://smutch.github.io/VersionControlTutorial/_images/vc-xkcd.jpg" width="400px"> .img-attr[Image: xkcd CC BY-NC 2.5 ] ] .pull-right[ ### Exhibit B <img src="http://www.phdcomics.com/comics/archive/phd101212s.gif" height="400px"> .img-attr[ Image: Jorge Cham www.phdcomics.com] ] --- # Git, Github & Rstudio #### Before: git only through the terminal 😢 -- *** ## Now: Rstudio + `usethis` 📦 == ❤️ `Git` & `GitHub` 🤩 .center[  ] --- # Configure git & GitHub ### Configure git **Check your configuration** ```r usethis::use_git_config() ``` **Set your configuration** Use your github username and and the email you used to sign-up on GitHub ```r usethis::use_git_config( user.name = "Jane", user.email = "jane@example.org") ``` --- # Configure GitHub authentication ### Get GITHUB Personal Authorisation Token ```r usethis::browse_github_pat() ``` <img src="assets/browse_github.png" height="300px"> --- ### Store in `.Renviron` file ```r usethis::edit_r_environ() ``` <img src="assets/GITHUB_PAT.png" height="400px"> --- # Initialise git ### Initialise **Rstudio project** with Git by **just checking a box!** It's now **a repository** <img src="assets/project_git.png" height="200px"> Forgot to? use `usethis::use_git()` --- # Git panel ## Integrated graphical user interface <br> .center[ <img src="assets/git_tab.png" height="300px"> ] --- # Git Rstudio workflow .pull-left[ #### view file status <img src="assets/git_view.png" height="150px"> #### stage files <img src="assets/git_add.png" height="150px"> ] .pull-right[ #### commit changes <img src="assets/git_commit.png" width="600px"> ] --- # Share on GitHub #### Create repo ```r usethis::use_github(protocol = "https") ``` <img src="assets/my_awesome_repo.png" width="500px"> #### Push further changes <img src="assets/push_github.png" height="50px"> --- # Anatomy of a GitHub Repo - **`README`**. Explain what your project is, and how to use it. + `usethis::use_readme_md()` + `usethis::use_readme_rmd()` - **`LICENSE`**. Without a licence, the contents of the repository are technically closed. + Examples licence [MIT](https://tldrlegal.com/license/mit-license): `usethis::use_mit_license(name = "Anna Krystalli")` + `?licenses`: details of functions available to generate licenses + [https://choosealicense.com/](https://choosealicense.com/) help on choosing a licence. - **`CONTRIBUTING.md`** - guidelines for contributors. + `usethis::use_tidy_contributing()` provides a realtively strict but instructive template - **`CODE_OF_CONDUCT.md`** set the tone for discourse between contributors. + `use_code_of_conduct()` --- # GitHub issues ### use GitHub issues to plan, record and discuss tasks. .pull-left[ #### issues <img src="assets/github_issues.png" width="600px"> ] .pull-right[ #### projects <img src="assets/github_projects.png" width="600px"> ] --- class: inverse, center, middle # Literate programming with Rmarkdown --- # Literate programming Programming paradigm first introduced by **Donald E. Knuth**. > ### Treat program as literature to be understandable to human beings > <br> > - focus on the logic and flow of human thought and understanding > - single document to integrate data analysis (executable code) with textual documentation, **linking data, code, and text** --- # Literate programming in R ### Rmarkdown (`.Rmd`) integrates: - a **documentantion** language (`.md`) - a **programming** language (`R`) - functionality to **"knit" them together** through 📦 `knitr` <br> ### features - ✅ provides a framework for writing narratives around code and data - ✅ Code re-run in a clean environment every time the document is "knit" --- background-image: url('assets/RMarkdownOutputFormats.png') background-size: contain # Rmarkdown outputs --- # Rmarkdown to html #### **File > New File > RMarkdown... > Document** .pull-left[  ] .pull-right.top[ <img src="assets/report_knit.png" width = 435px> ] --- # Applications in research ### Rmd documents can be useful for a number of research related [long form documentation](http://r-pkgs.had.co.nz/vignettes.html) materials: .pull-left[ <br> - Documentation of code & data (eg 📦 [DataMaid](https://github.com/ekstroem/dataMaid)) - Electronic Notebooks - Supplementary materials - Reports - Papers ] .pull-right[  ] --- # Rmd Vs Word #### Spell check in Rstudio! <img src="assets/spell_check.png" height=40px> ## 📦 [redoc](https://github.com/noamross/redoc) <small>**HOT OFF THE PRESS**</small> **Enables a two-way R Markdown-Microsoft Word workflow**. It generates Word documents that can be de-rendered back into R Markdown, **retaining edits on the Word document**, including tracked changes.  --- # Publish to the web for free! - **RPubs**: Publish rendered rmarkdown documents on the web with the click of a button <http://rpubs.com/> - **GitHub**: Host your site through [`gh-pages`](https://pages.github.com/) on GitHub. Enable in GitHub repo ⚙️**Settings** .center[ <img src="assets/gh-pages.png" height=400px> ] --- class: inverse, center, middle # Rmarkdown extensions Many great packages and applications build on rmarkdown. All this makes it [incredibly versatile](https://rmarkdown.rstudio.com/gallery.html). --- # [bookdown](https://bookdown.org/yihui/bookdown/) #### Create and mantain online books Authoring with R Markdown. Offers: - cross-references, - citations, - HTML widgets and Shiny apps, - tables of content and section numbering The publication can be exported to HTML, PDF, and e-books (e.g. EPUB) ### Examples - [rOpenSci Software Review policies](https://ropensci.github.io/dev_guide/) - [Geocomputation in R](https://geocompr.robinlovelace.net/) ### [Thesisdown](https://github.com/ismayc/thesisdown) An updated R Markdown thesis template using the bookdown package --- # [pkgdown](http://pkgdown.r-lib.org/articles/pkgdown.html) #### For buidling package documentation Produce **function references** from `.Rd` files and **demonstrate function use** through long form documentation (vignettes).  --- # [workflowr](https://jdblischak.github.io/workflowr/) pkg #### Build analyses websites and organise your project Makes it easier for researchers to organize projects and share results. Includes **checks to ensure rendered versions correspond to up to date versions of code**. .pull-left[ 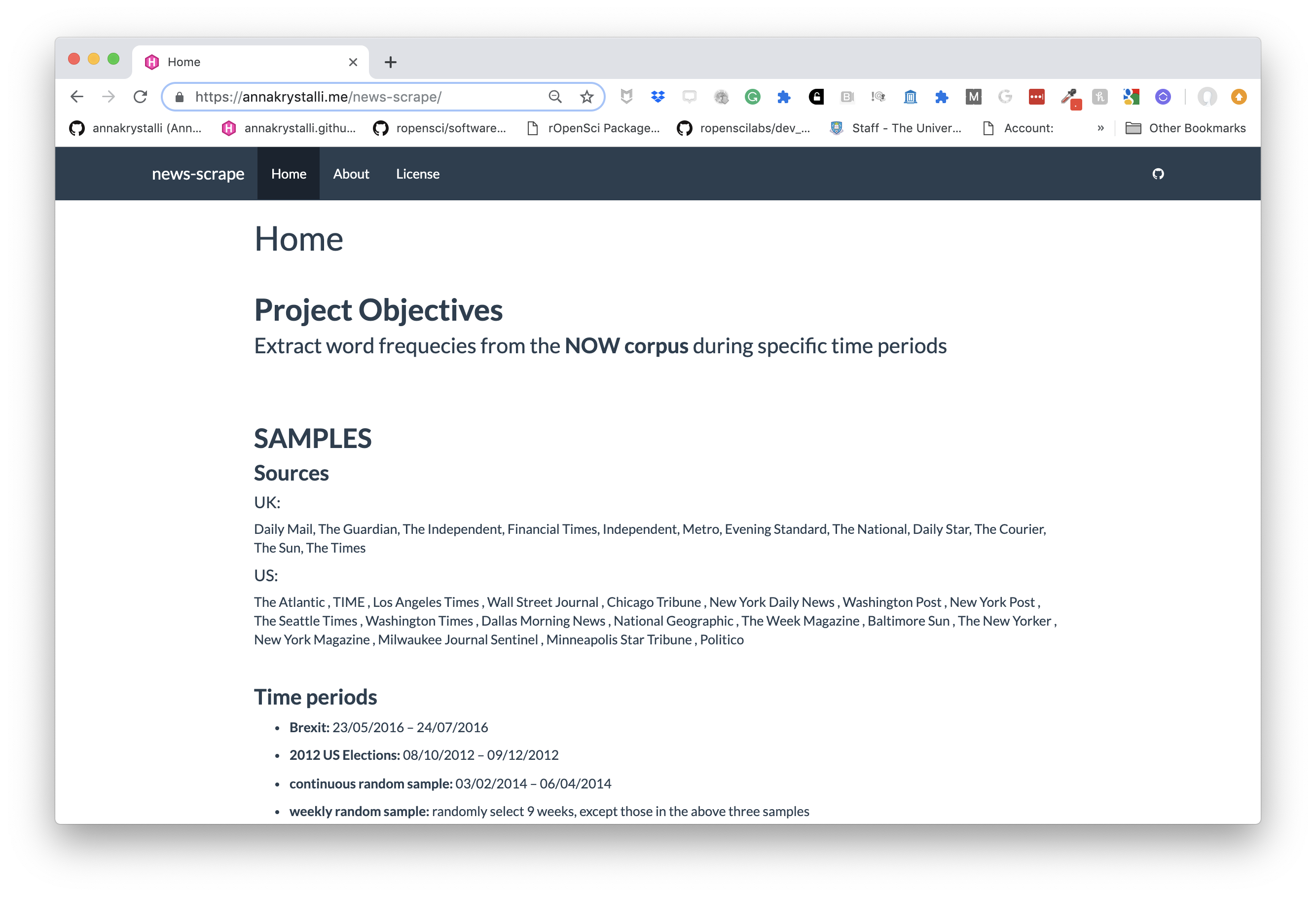 ] .pull-right[ 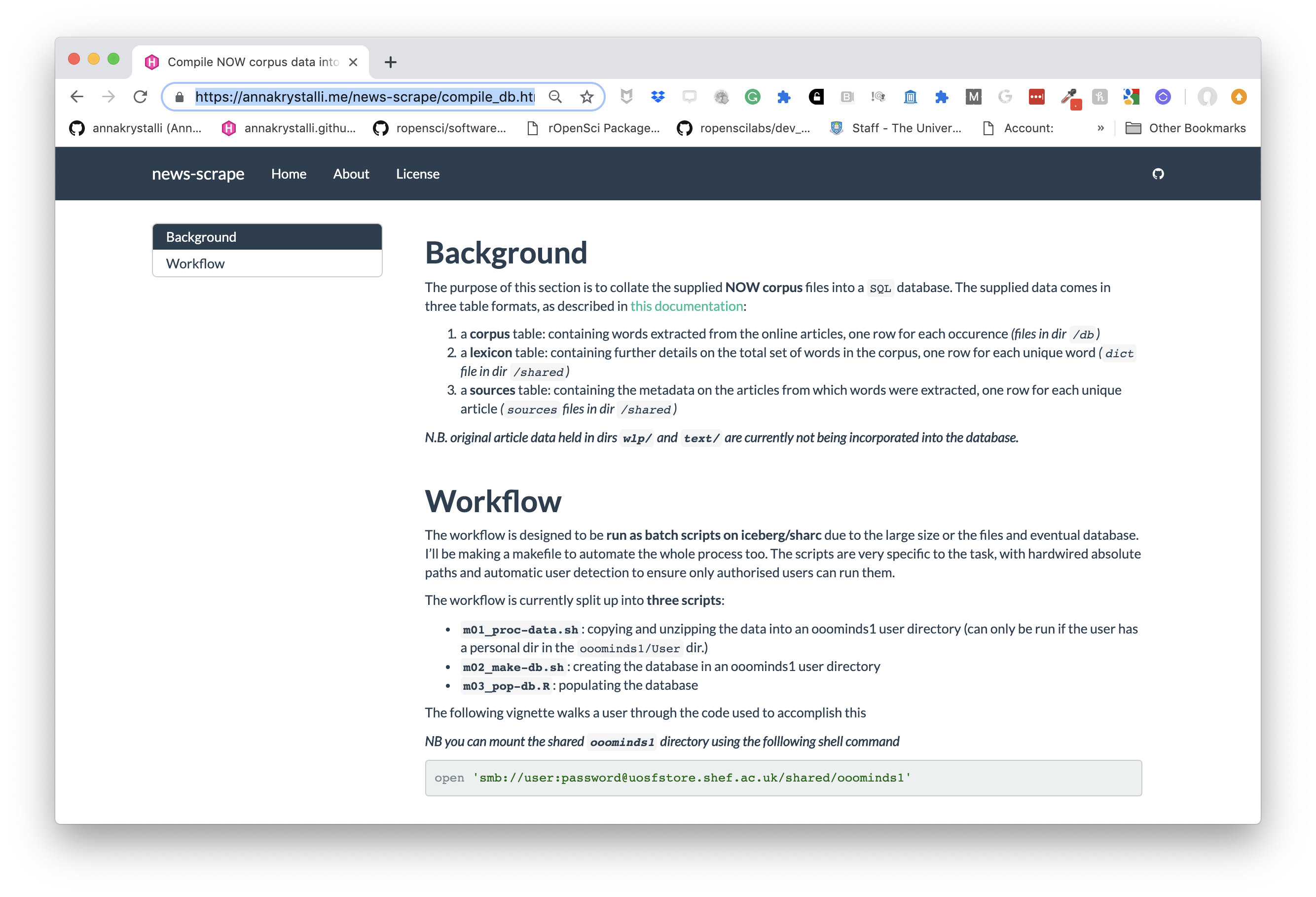 ] --- # [blogdown](https://bookdown.org/yihui/blogdown/) ## For creating and mantaining blogs through R. Check out <https://awesome-blogdown.com/>, **a curated list of awesome #rstats blogs in blogdown** for inspiration! [](https://masalmon.eu/) --- # presentations ## A number of existing frameworks ### [xaringan](https://github.com/yihui/xaringan) 📦 Presentation Ninja 幻灯忍者 · 写轮眼 .center[ [<img src='assets/xaringan.png' height=350px>](https://slides.yihui.name/xaringan/#1) ] --- class: inverse, center, middle # Managing code --- # Managing analysis code ## Separate function definition and application .pull-left[ <br> - When a project is new and shiny, an **analysis script usually contains many lines of directly executated code.** - As it matures, **reusable chunks get pulled into their own functions**. - The actual analysis scripts then become relatively short, and **functions defined in separate R scripts.** ] .pull-right[  ] --- # R Package Structure ## Used to share functionality with the R community - Useful **conventions** - Useful **software development tools** - Easy **publishing** through GitHub <br> .center[ <img src="assets/package_friends.png" height=250px> ] --- # Build panel ## Integrated graphical user interface <br> .center[ <img src="assets/build_panel.png" height="300px"> ] --- # R Package conventions: - **metadata**: in the **`DESCRIPTION` file** - **functions** in **`.R` scripts** in the **`R/` folder** - **tests** in the **`tests/` folder** - **Documentation:** - _functions_ using **Roxygen notation** - _workflows_ using **`.Rmd` documents** in the **`vignettes/`** folder --- # `DESCRIPTION` file #### Package metadata ``` Package: gaitr Type: Package Title: Functions to support BMC gait analysis Description: Helpers to analyse processed gait data. Version: 0.0.9000 Authors@R: c(person(given = "Anna", family = "Krystalli", role = c("aut", "cre"), email = "annakrystalli@googlemail.com"), person(given = "Lorenza", family = "Angelini", role = "aut", email = "l.angelini@sheffield.ac.uk")) License: MIT + file LICENSE ``` --- # citation ```r citation("gaitr") ``` ``` ## ## To cite package 'gaitr' in publications use: ## ## Anna Krystalli and Lorenza Angelini (2019). gaitr: Functions to ## support BMC gait analysis. R package version 0.1.1. ## https://github.com/annakrystalli/gaitr ## ## A BibTeX entry for LaTeX users is ## ## @Manual{, ## title = {gaitr: Functions to support BMC gait analysis}, ## author = {Anna Krystalli and Lorenza Angelini}, ## year = {2019}, ## note = {R package version 0.1.1}, ## url = {https://github.com/annakrystalli/gaitr}, ## } ``` --- # Dependency management It’s the job of the `DESCRIPTION` file to **list the packages that your code depends on**. ``` Imports: dplyr, purrr, here, broom, tibble, magrittr, janitor, ggplot2 Suggests: knitr, rmarkdown ``` #### add dependency ```r usethis::use_package("forcats", type = "Imports") ``` --- # Functions in `R/` ### example function script Create a new function `.R` file in the `R/` folder ```r usethis::use_r("add") ``` ``` R └── add.R 0 directories, 1 files ``` --- # Document functions with `Roxygen` - Create help files on build (autogenerated `.Rd` files in `man/`) - Specify which functions are exported (autogenerated `NAMESPACE`) ```r #' Add together two numbers. #' #' @param x A number. #' @param y A number. #' @return The sum of x and y. #' @examples #' add(1, 1) #' add(10, 1) add <- function(x, y) { x + y } ``` --- # [tests](http://r-pkgs.had.co.nz/tests.html) ## Tests provide confidence in what the code is doing. .center[  ] --- # Example test ```r usethis::use_test("add") ``` Creates a `tests/` folder with the following files ``` tests ├── testthat │ ├── test-add.R └── testthat.R ``` ##### test-add.R ```r context("test-add") test_that("add works", { expect_equal(add(2, 2), 4) }) ``` --- # Continuous Integration ## A cloud testing framework for automating your tests - Monitor the effect of changes to the code - Safe onboarding of contributions .pull-left[ ### Start with a `.travis.yml` file ```r usethis::use_travis() ``` ] .pull-right.center[ <img src="assets/travis.png" height=250px> ] --- #### Resulting `.travis.yml` file template ``` language: R sudo: false cache: packages ``` #### instructions to enable TRAVIS CI ``` ✔ Writing '.travis.yml' ✔ Adding '^\\.travis\\.yml$' to '.Rbuildignore' ● Turn on travis for your repo at https://travis-ci.org/profile/annakrystalli ● Copy and paste the following lines into '/Users/Anna/Documents/workflows/talks/README.md': <!-- badges: start --> [](https://travis-ci.org/annakrystalli/talks) <!-- badges: end --> ``` --- class: inverse, center, middle # Research compendia --- # A Research compendium ### The paper is the advertisement > “an article about computational result is advertising, not scholarship. The **actual scholarship is the full software environment, code and data, that produced the result.**” *John Claerbout paraphrased in [Buckheit and Donoho (1995)](https://statweb.stanford.edu/~wavelab/Wavelab_850/wavelab.pdf)* ### The concept of a Research Compendium >“ ...We introduce the **concept of a compendium** as both a **container for the different elements** that make up the document and its computations (i.e. text, code, data, ...), and as a **means for distributing, managing and updating the collection**." [_Gentleman and Temple Lang, 2004_](https://biostats.bepress.com/bioconductor/paper2/) --- # Research compendia in R .pull-left[ 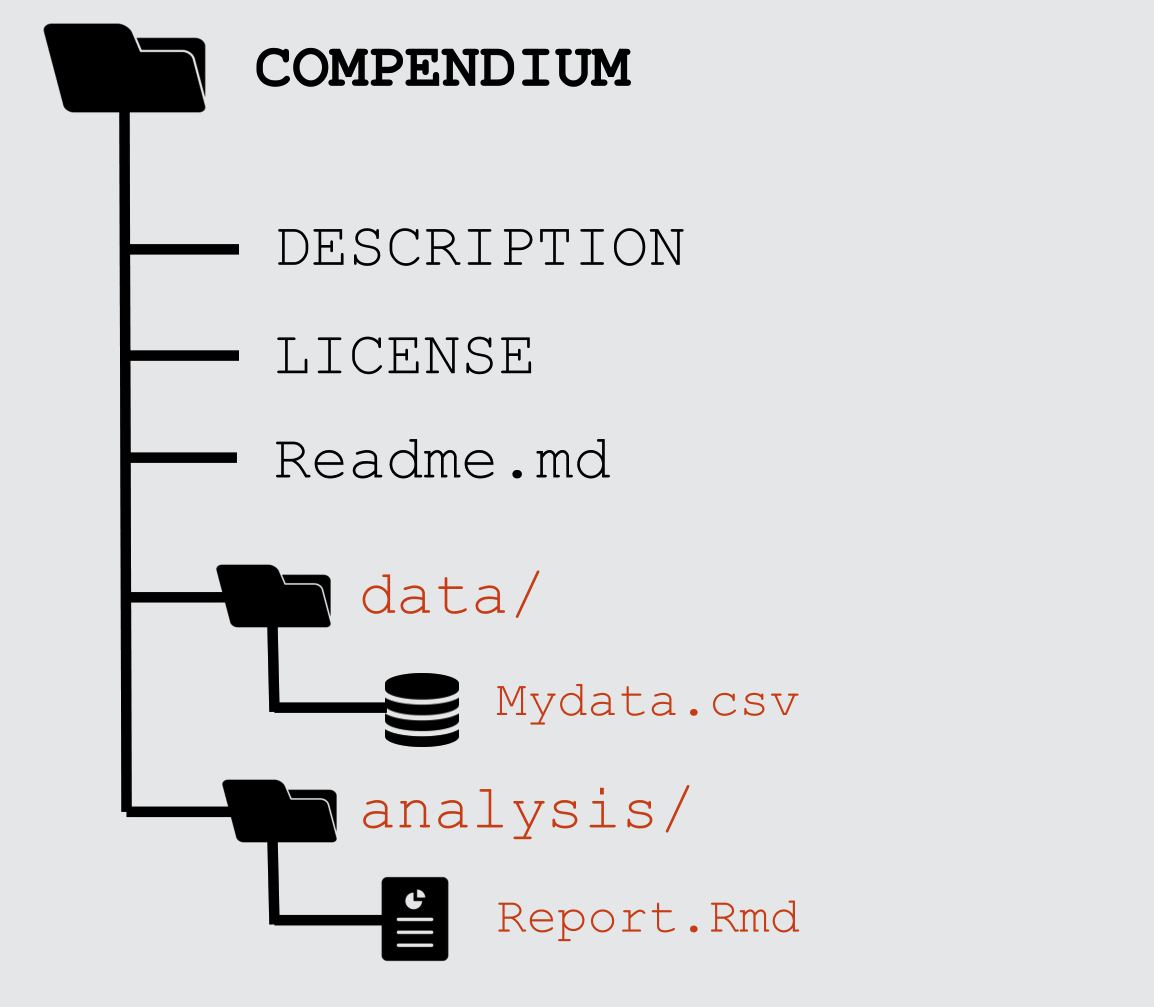 ] .pull-right[  ] .img-attr[slides: [_Karthik Ram: rstudio::conf 2019 talk_](https://github.com/karthik/rstudio2019)] <br> **Ben Marwick, Carl Boettiger & Lincoln Mullen (2018)** [_Packaging Data Analytical Work Reproducibly Using R (and Friends)_](https://peerj.com/preprints/3192/) --- # Example compendium .pull-left[ **Paper**: ##### Boettiger, C. (2018) *From noise to knowledge: how randomness generates novel phenomena and reveals information*. <https://doi.org/10.1111/ele.13085> <img src="assets/Boettiger-2018.png" heigth="250px" width="400px"> ] .pull-right[ **Compendium** ##### *cboettig/noise-phenomena: Supplement to: "From noise to knowledge: how randomness generates novel phenomena and reveals information"* http://doi.org/10.5281/zenodo.1219780 <img src="assets/boettiger_compendium.png" heigth="250px" width="400px"> ] --- # `rrtools`: Creating Compendia in R ### "The goal of rrtools is to provide **instructions, templates, and functions** for making a **basic compendium** suitable for writing **reproducible research with R**." <br> -- ### Install [`rrtools`](https://github.com/benmarwick/rrtools) from GitHub ```r # install.packages("devtools") devtools::install_github("benmarwick/rrtools") ``` --- # Create compendium ```r rrtools::create_compendium("~/Documents/workflows/rrcompendium") ``` ``` ✔ Setting active project to '/Users/Anna/Documents/workflows/rrcompendium' ✔ Creating 'R/' ✔ Creating 'man/' ✔ Writing 'DESCRIPTION' ✔ Writing 'NAMESPACE' ✔ Writing 'rrcompendium.Rproj' ✔ Adding '.Rproj.user' to '.gitignore' ✔ Adding '^rrcompendium\\.Rproj$', '^\\.Rproj\\.user$' to '.Rbuildignore' ✔ Opening new project 'rrcompendium' in RStudio ✔ The package rrcompendium has been created ✔ Opening the new compendium in a new RStudio session... Next, you need to: ↓ ↓ ↓ ● Edit the DESCRIPTION file ● Use other 'rrtools' functions to add components to the compendium ``` --- # Prepare for GitHub ```r rrtools::use_readme_rmd() ``` .pull-left[ ``` ✔ Creating 'README.Rmd' from template. ✔ Adding 'README.Rmd' to `.Rbuildignore`. ● Modify 'README.Rmd' ✔ Rendering README.Rmd to README.md for GitHub. ✔ Adding code of conduct. ✔ Creating 'CONDUCT.md' from template. ✔ Adding 'CONDUCT.md' to `.Rbuildignore`. ✔ Adding instructions to contributors. ✔ Creating 'CONTRIBUTING.md' from template. ✔ Adding 'CONTRIBUTING.md' to `.Rbuildignore`. ``` ] .pull-right[  ] --- # Create analysis folder ```r rrtools::use_analysis() ``` ``` ✔ Adding bookdown to Imports ✔ Creating 'analysis' directory and contents ✔ Creating 'analysis' ✔ Creating 'analysis/paper' ✔ Creating 'analysis/figures' ✔ Creating 'analysis/templates' ✔ Creating 'analysis/data' ✔ Creating 'analysis/data/raw_data' ✔ Creating 'analysis/data/derived_data' ✔ Creating 'references.bib' from template. ✔ Creating 'paper.Rmd' from template. Next, you need to: ↓ ↓ ↓ ↓ ● Write your article/report/thesis, start at the paper.Rmd file ● Add the citation style library file (csl) to replace the default provided here, see https://github.com/citation-style-language/ ● Add bibliographic details of cited items to the 'references.bib' file ● For adding captions & cross-referencing in an Rmd, see https://bookdown.org/yihui/bookdown/ ● For adding citations & reference lists in an Rmd, see http://rmarkdown.rstudio.com/authoring_bibliographies_and_citations.html ``` --- # `paper.Rmd` to `paper.pdf` .pull-left[ **Rmd** <img src="assets/paper_rmd.png" > ] .pull-right[ **pdf** <img src="assets/paper_pdf.png" > ] --- # Capturing dependencies ```r rrtools::add_dependencies_to_description() ``` ``` Imports: bookdown, ggplot2 (>= 3.0.0), ggthemes (>= 3.5.0), here (>= 0.1), knitr (>= 1.20), rticles (>= 0.6) ``` --- # Further Helpers ## 📦 `rticles` Contains a **suite of custom R Markdown templates for popular journals**, simplifying the creation of documents that conform to research paper submission standards. --- # 📦 `citr` RStudio Add-in to **Insert Markdown Citations** <img src="assets/citr-insert.png" width="700px"> --- background-image: url('assets/reproducible-data-analysis_042.png') background-size: contain # Succesful Reproducibility <br> <br> <br> <br> <br> <br> <br> <br> <br> <br> <br> <br> <br> <br> .bottom[ .img-attr[slide: [_Karthik Ram: rstudio::conf 2019 talk_](https://github.com/karthik/rstudio2019)] ] --- class: inverse, center, middle # Questions? --- # Further reading .pull-left[ #### Version Control - [Happy Git and GitHub for the useR](https://happygitwithr.com/) #### RMarkdown - [R Markdown: The Definitive guide](https://bookdown.org/yihui/rmarkdown/) - [RMarkdown Driven Development (RmdDD)](https://emilyriederer.netlify.com/post/rmarkdown-driven-development/): Blog post by Emily Riederer #### R Packages - [R packages](https://r-pkgs.org/) by Hadley Wickham and Jenny Bryan ] .pull-right[ #### Research Compendia - Karthik Ram: [_rstudio::conf 2019 talk_](https://github.com/karthik/rstudio2019) #### Tutorials - [Rstudio Essentials](https://resources.rstudio.com/) Webinar series - [rrresearch](https://annakrystalli.me/rrresearch/): ACCE DTP course on Research Data & Project Management ]